PSMD (Peak width of Skeletonized Mean Diffusivity)
PSMD is a robust, fully-automated and easy-to-implement marker for cerebral small vessel disease based on diffusion tensor imaging, white matter tract skeletonization (as implemented in FSL-TBSS) and histogram analysis. The software package provided through this page allows to calculate PSMD from diffusion tensor imaging data for research use.
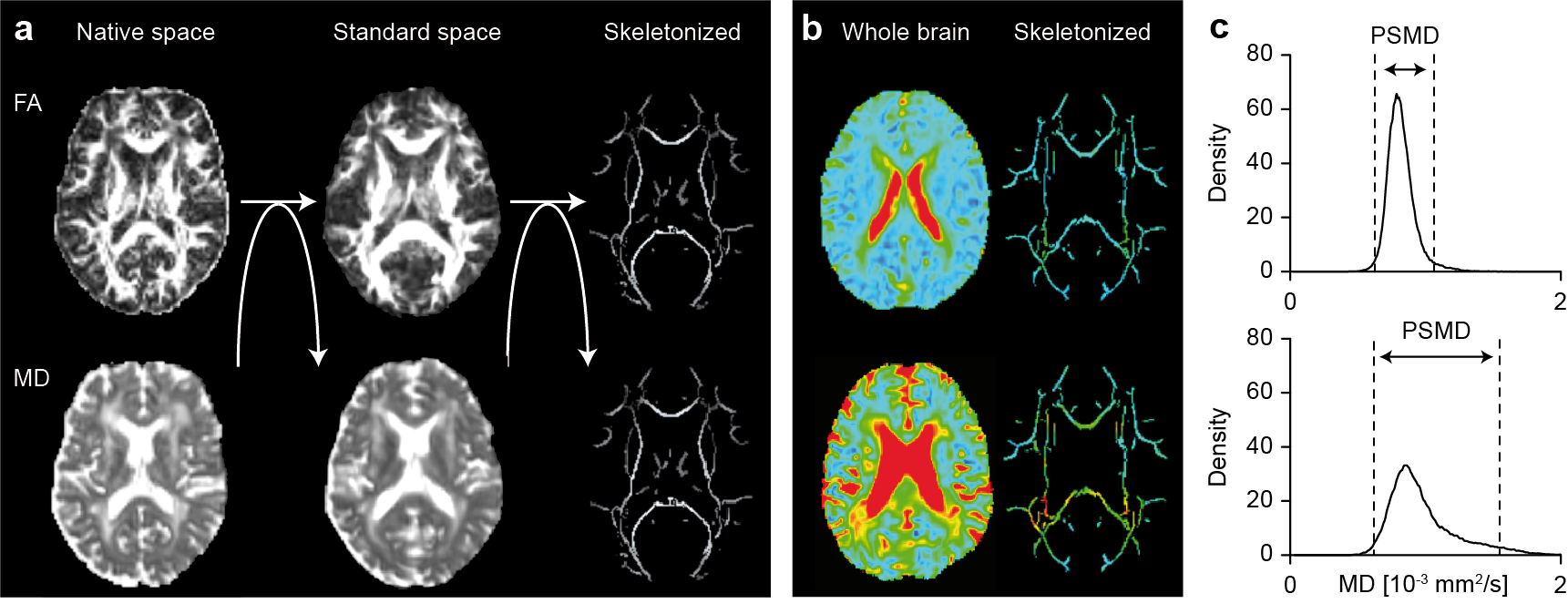
Figure: Calculation of PSMD. a) MD images are first skeletonized using the FA image and the FSL-TBSS pipeline. b) Example images of a mildly (top) and severely affected (bottom) patient. c) Histrogram analysis on the skeletonized data. PSMD quantifies the peak width of the histogram. The same two patients as in b are shown. Figure modified from Baykara et al., Annals of Neurology 2016.
For more background information, please see the paper published in Annals of Neurology.
Please note that PSMD is not a medical device and for research use only. Any clinical use (e.g. for diagnosis, prognosis or monitoring) is prohibited by law (see e.g. medical device regulation in the EU).
PSMD is under constant development for new features and bug fixes. Please see the development repository for more details and a roadmap for future features.